Biobanks advance precision medicine research through collaboration
Access powerful biobank data for your own projects or contribute to a biobank
Population proteomics drives innovative precision medicine research worldwide
Almost 225,000
samples
To date, nearly a quarter million samples have been run by consortium members on the SomaScan Assay.
With advances in high-plex proteomic technologies, the ability to profile thousands of proteins at once with high specificity and sensitivity has opened the door to working with larger scale, population sized cohorts. These types of population proteomics studies are generating new datasets that enable a more comprehensive overview of the dynamics of protein expression in a wide range of biological samples such as plasma, serum, urine and CSF, revealing new insights into gene-to-protein correlations, novel biology, mechanisms of action and new therapeutic targets.
As curated sample repositories, biobanks serve as a key resource for these studies, providing valuable samples and related data in the thousands that can be translated to population proteomics approaches. Additionally, longitudinal sampling and collaborations across projects quickly increases the value of what biobanks can offer. With the large-scale outputs that can be produced using the SomaScan assay, many biobanks maintain this protein profiling data on larger sample cohorts for qualified researchers to leverage.
Accelerate your own research by accessing data already discovered by biobanks using the SomaScan Assay
This table is updated quarterly with new data about biobanks conducting innovative population-based proteomics research. To learn more about the biobanks, please click “View now” next to each.
Resources for biobanks: How to include SomaScan Assay data in your research
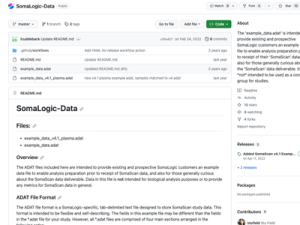
The ADAT file format is a Standard BioTools-specific, tab-delimited text file designed to store Somascan study data. This format is intended to be flexible and self-explanatory. When sample analysis is completed, Standard BioTools normalizes the data to ensure data quality and then creates an ADAT file for delivery of results to the researcher.
If you have questions or need assistance with ADAT files, please email Standard BioTools Technical Support here.

A data repository serves as a structured digital storage facility, often resembling databases or data lakes, where research data and analytical resources are systematically organized and made accessible in accordance with FAIR (Findable, Accessible, Interoperable, and Reusable) standards. Data repositories exist in two primary forms: open access and closed access.
Learn why researchers are increasingly encouraged – or required – to upload data to a repository before publishing their findings.
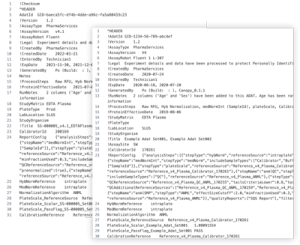
Data repositories can exist in two primary forms: open access, which are open to the public, or closed access, restricted to specific users or groups, and they may cater to discipline-specific or broader research datasets. Choosing which type of data repository to upload to depends on several factors. See the table below for details:
Researchers can analyze and visualize all their studies in one convenient view on the Standard BioTools Life Sciences Portal. Digital resources, such as DataDelve™ Statistics and the SomaScan Menu, are easily accessed from the portal, too.
The Standard BioTools Technical Support team is always available to support researchers and repository owners. Email us with questions.

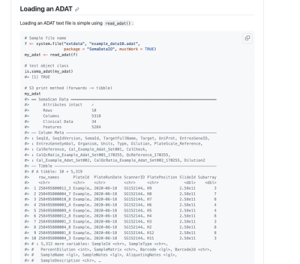
SomaScan data are presented in an ADAT format, a Standard BioTools-specific, tab-delimited text file that’s small enough to fit on a standard desktop computer. ADAT files can be read easily into any R/Python scripting languages using our ADAT reader function in SomaDataIO (R package) or Canopy (Python package). For uploading to public data repositories, we recommend that SomaScan data owners provide ADAT files formatted as they received them from Standard BioTools.
For more information on ADAT format or handling SomaScan data, please email the Standard BioTools Technical Support team.
![]()
The Standard BioTools Technical Support team is available throughout the analysis process – from initial questions and sample prep, to bioinformatics analysis and questions and final delivery of data.
Please email our team here.
Interested in joining the SomaScan Proteomics Consortium?
The SomaScan Proteomics Consortium was founded by Russell Bowler, MD, PhD, to enable researchers to increase the power of their studies by leveraging high-plex proteomic profiling with the SomaScan® Assay across diverse populations worldwide, for more powerful biomarker discovery and deeper insights into human biology. Members are committed to sharing data for the purpose of increasing efficiency and efficacy, as well as for fostering discussions about the development and pursuit of new studies and research directions.
For general inquiries or to join the consortium, please contact
Russell Bowler, MD, PhD.




