Biomarker discovery leading the way in PD-L1 expression and PD-1 checkpoint inhibitor therapy
Investigate the potential of targeting PD-L1 expression & PD-1 checkpoint pathway in cancer.
Biomarker discovery leading the way in PD-L1 expression and PD-1 checkpoint inhibitor therapy
Immunotherapy is a relatively new type of cancer treatment. Unlike chemotherapy, surgery, and radiation that target the cancer itself, immunotherapies are designed to stimulate the immune response to aid in destroying cancer cells.
In doing so, immunotherapy has been a game-changing addition to the cancer treatment toolbox and has several advantages over traditional radiation and chemotherapy treatments, such as:
- Longer-lasting benefits after completion of treatment1
- Better response in advanced cancers1
- Fewer severe side effects2
Immune checkpoint blockade is a type of immunotherapy that works by disrupting normal T-cell surveillance on our cells. T-cells are an important part of our adaptive immune system that become trained to recognize foreign molecules (viruses, pathogens, bacteria) and quickly fight them the next time we are exposed. (This is how many vaccines work.)
What is the PD-1 and PD-L1 pathway?
Our immune cells have a protein called Programmed Cell Death Ligand 1 (PD-L1) on their surface which binds to the Programmed Cell Death Protein (PD-1) receptor on T-cells and prevents T-cells from attacking healthy cells. This interaction, PD-1 binding to PD-L1, is called a checkpoint: it is a check on the action of T-cells, preventing them from destroying healthy cells.
The PD-L1 checkpoint in cancer treatment
Some cancer cells express PD-L1 and therefore evade attack by T-cells. Upon binding PD-1, the immune response will be inactivated allowing the cancer cell to continue maturing. Checkpoint inhibitor drugs interrupt this communication between PD-1 and PD-L1, which removes the brakes from the immune system and allows T-cells to target and destroy cancer cells.
While checkpoint inhibitors have revolutionized cancer treatment, their efficacy varies widely among patients, and they can have their own side-effects. (i.e. T-cells kill healthy cells too.) They’re also expensive and do not necessarily work for everyone or for all cancer types.
Identifying individuals who are most likely to benefit from these therapies is crucial for optimizing treatment outcomes and minimizing adverse effects. This is where the discovery and utilization of biomarkers come into play, offering a promising avenue for predicting patient response to checkpoint inhibitor therapy.
Predicting response to checkpoint inhibitor therapy
Identifying biomarkers to indicate responders vs. non-responders is a critical part of the treatment process3 yet no clear set of biomarkers exist that can reliably identify those who will respond favorably to immunotherapy.4 It is clear that genetic factors alone, such as variations in human leukocyte antigen (HLA) class I molecules, are not consistently correlated with survival among cohorts of patients on checkpoint inhibitor therapy.5
Examining the tumor itself is currently the best way to predict an individual’s response to checkpoint inhibitor therapy. Tumor PD-L1 expression, tumor mismatch repair deficiency (a deficiency in DNA repair mechanism which leads to hypermutation), and tumor mutational burden (the number of mutations within the tumor DNA) are all characteristics that help predict tumor response to therapy.6 However, a central limitation of this method is the need for tissue biopsy—an invasive process with risks of its own.
Biomarker identification with the SomaScan® 11K Assay
But, what if there was a way to identify individuals who will respond favorably to checkpoint inhibitor therapy based on biomarkers found in blood?
The SomaScan Platform is an aptamer-based high-plex, high-throughput protein profiling technology which allows comprehensive analysis of circulating proteins in blood samples. The newest version of the assay now includes coverage of an unprecedented 11,000 protein measurements – that’s half of the human proteome. Using proteins included on the SomaScan 11K Menu as a framework, we can overlay pathway analysis tools such as STRING (Search Tool for the Retrieval of Interacting Genes/Proteins), to highlight all proteins in the PD-1/PD-L1 pathway that can be measured by SomaScan. Reliable measurement of broad networks of proteins is a key first step in identifying and testing potential biomarkers of therapy response.
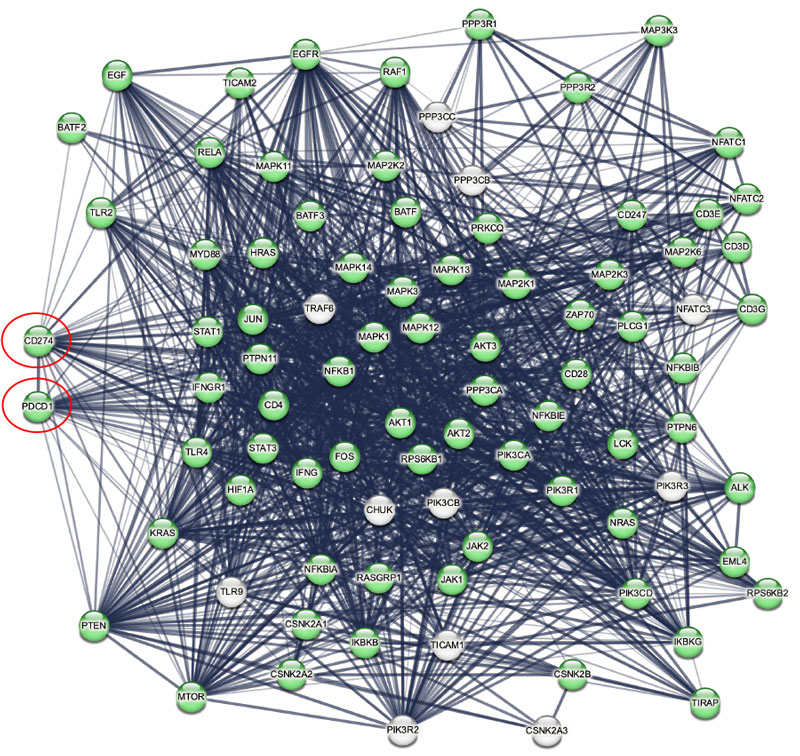
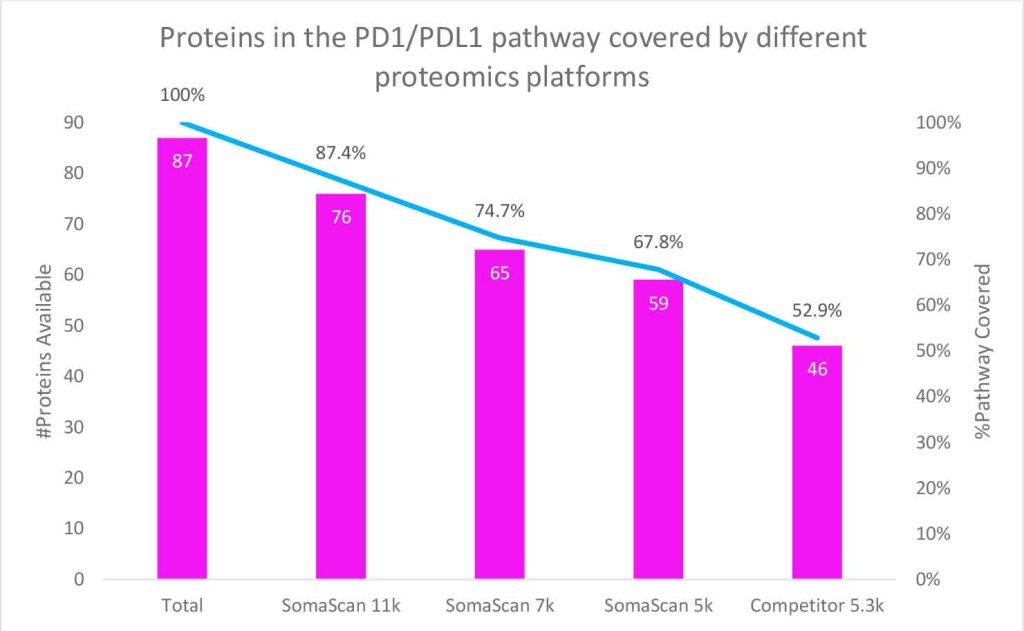
In this figure, STRING network mapping shows 87 protein interactions in the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway “PD-L1 Expression and PD-1 Checkpoint Pathway in Cancer”.7 PD-1 (PDCD1) and PD-L1 (CD274) are circled in red; proteins in green (for example CD28, IFNg, Tyrosine-protein kinase JAK1 and JAK2, and Signal transducer and activator of transcription 1 and 3 (STAT1, STAT3)) (n=76 or 87% of the pathway) are available on the SomaScan 11K Menu.
When compared, previous versions of the SomaScan Assay (featuring 5k and 7k analyte menus) provided 68% and 75% coverage of the network respectively. This surpasses a competing platform that has only 53% coverage. This highlights how the expanded coverage offered by the SomaScan 11K Assay significantly enhances the potential for biomarker discovery.
Researchers utilizing the SomaScan Assay have been leading the way in blood-based biomarker discovery for checkpoint inhibitor therapy response. The technology has been used to identify biomarkers of toxicity and outcome in patients receiving checkpoint inhibitor therapy in metastatic melanoma8 and non-small cell lung cancer.9,10
Read our whitepaper for more examples of how the SomaScan Platform and circulating protein biomarkers are a promising avenue for predicting patient response to cancer immunotherapies.

Go further with mass cytometry
SomaLogic, now part of Standard BioTools™, combines a broad discovery-enabled platform with a suite of high-plex imaging instruments. The Hyperion XTi™ instrument is an exciting avenue to pursue the complementary relationship between blood-based proteomics and imaging mass cytometry. In oncology research in particular, it can be used to further define biomarkers for use in prognosis and understanding the mechanism of action of how cancer cells evade the immune system.
Making biomarker-driven cancer care possible
Novel immunotherapies are a turning point in cancer treatment, enabling oncologists to combine precision medicine with broader treatment modalities. Stratifying individuals based on their anticipated response to checkpoint inhibitor therapies promises to streamline treatment, saving valuable time, resources, and lives.
Central to this framework lies a dependable set of biomarkers that remain unaffected by tumor heterogeneity, are responsive and indicative of tumor evolution, characterize the tumor microenvironment, and can be measured with minimal patient burden.
The SomaScan Assay’s comprehensive coverage of the human proteome presents new potential for discovering precise, sensitive, and low-abundance plasma biomarkers for this use. With the ability to simultaneously detect approximately 11,000 proteins across a 10-log dynamic range from just 55 µl of blood, high-plex, high-throughput biomarker profiling holds the key to unlocking transformative insights in therapeutic development.
Additional resources
References
- Borcoman E, Kanjanapan Y, Champiat S, Kato S, Servois V, Kurzrock R, Goel S, Bedard P, Le Tourneau C. Novel patterns of response under immunotherapy. Ann Oncol. 2019 Mar 1;30(3):385-396.
- Hofman P. Is the onset of adverse effects of immunotherapy always bad news for the patients…?-certainly not! Ann Transl Med. 2019 Mar;7(Suppl 1):S5.
- Li H, van der Merwe PA, Sivakumar S. Biomarkers of response to PD-1 pathway blockade. Br J Cancer. 2022;126(12):1663-1675.
- Pilard C, Ancion M, Delvenne P, Jerusalem G, Hubert P, Herfs M. Cancer immunotherapy: it’s time to better predict patients’ response. Br J Cancer. 2021;125(7):927-938.
- Negrao MV, Lam VK, Reuben A, et al. PD-L1 Expression, Tumor Mutational Burden, and Cancer Gene Mutations Are Stronger Predictors of Benefit from Immune Checkpoint Blockade than HLA Class I Genotype in Non-Small Cell Lung Cancer. J Thorac Oncol. 2019;14(6):1021-1031.
- Hanna NH, Schneider BJ, Temin S, et al. Therapy for Stage IV Non-Small-Cell Lung Cancer Without Driver Alterations: ASCO and OH (CCO) Joint Guideline Update. J Clin Oncol. 2020;38(14):1608-1632.
- Szklarczyk D, Kirsch R, Koutrouli M, et al. The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023;51(D1):D638-D646.
- Mooradian M, Gu X, Lawrence DP, et al. Predictive plasma proteomic biomarkers of immunotherapy toxicity in patients (pts) with metastatic melanoma (MM). Journal of Clinical Oncology. 2018;36(15_suppl):e21569-e21569.
- Bar J, Leibowitz R, Reinmuth N, et al. Biological insights from plasma proteomics of non-small cell lung cancer patients treated with immunotherapy. bioRxiv. 2024:2024.2001.2002.573876.
- Naidoo J, Reinmuth N, Puzanov I, et al. 1229 Pre-treatment plasma proteomics-based predictive biomarkers for immune related adverse events in non-small cell lung cancer. Journal for ImmunoTherapy of Cancer. 2023;11(Suppl 1):A1356-A1356.
- Le Rochais M, Hemon P, Pers JO, Uguen A. Application of High-Throughput Imaging Mass Cytometry Hyperion in Cancer Research. Front Immunol. 2022;13:859414.
More blogs
BlogHow Blood Proteomics Is Accelerating Neuro Drug Discovery
Explore how researchers are using blood-based proteomics – including data from the Global Neurodegeneration Proteomics Consortium (GNPC) – to uncover new protein signatures, genetic risk associations and mechanistic insights into aging and neurodegeneration.
BlogGNPC Neurodegeneration Proteomics Dataset
On July 15, 2025, the Global Neurodegeneration Proteomics Consortium (GNPC) published four open‑access papers unveiling the largest disease‑specific proteomic resource ever assembled.
BlogIndependent study validates the SomaScan Assay as the most precise and comprehensive plasma proteomic platform
A recent independent study by Alkahest, published as a preprint on bioRxiv1, compared the technical precision and performance of multiple plasma proteomic platforms using a common set of samples.





