The ability to profile thousands of proteins in a single assay begins with exquisite specificity
See the exquisite specificity of the SomaScan® Assay
The SomaScan [Assay] also has a two-step approach to specificity that is sometimes forgotten by non-experts…between these two steps, the assay turns out to be quite highly specific.”
Professor of Medicine,
University of California in San Francisco
Why modified aptamers?
The SomaScan Platform uses unique modified aptamers — short sequences of DNA — as protein affinity reagents. Our proprietary SOMAmer® (Slow Off-Rate Modified Aptamer) Reagents are chemically modified aptamers to achieve specificity and sensitivity.
Shape specificity
By using uniquely modified DNA with hydrophobic functional groups, our SOMAmer Reagents achieve a greater degree of shape matching, enabling discernment between nearly identical proteins.
Slow off-rates
SOMAmer Reagents are selected for slow off-rates when correctly bound to their intended target to ensure that correct proteins remain preferentially bound during rigorous washing.
Polyanionic competition
Polyanionic competitors are used to universally prevent rebinding of transient, non-specific interactions. There is no such universal competitor for antibodies.
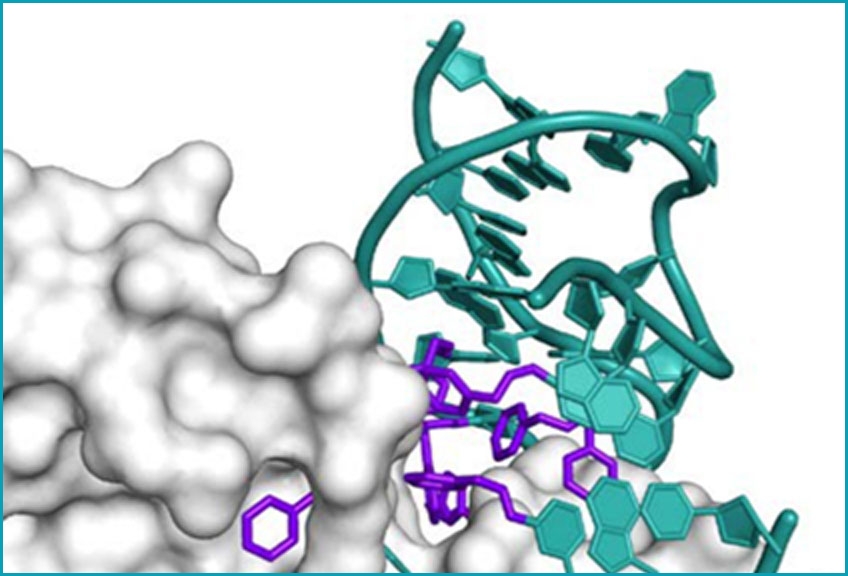
X-ray crystal structure of a
SOMAmer Reagent bound
to PDGF-BB
Modifications to the bases are shown in purple, and the DNA backbone and unmodified bases are in teal.
Typical dissociation
for a
SOMAmer Reagent
This graph shows the typical dissociation for a SOMAmer Reagent over time, in this case targeting C3.

How do aptamers
compare to antibodies?
Traditionally, affinity-based platforms have used antibodies as reagents. But SomaLogic pioneered the use of aptamers more than 20 years ago to address the challenges researchers face in using antibodies, including performance and natural variability in structure. Download a quick comparison here.
Primary validation and orthogonal confirmation of SOMAmer Reagent specificity
SOMAmer Reagents are protein-binding affinity reagents targeted to a specific protein.
Primary validation of all SOMAmer Reagents
- Determination of the equilibrium binding affinity dissociation constant (KD)
- Pulldown assay of cognate protein in buffer using the SOMAmer Reagent
- Demonstration of buffer dose response in the SomaScan Assay
- Estimation of endogenous signal in plasma
Orthogonal confirmation
- At least 66% of SOMAmer Reagents have at least one additional source of orthogonal confirmation
- Specificity of the SOMAmer Reagent for its cognate protein has been confirmed by orthogonal methods, such as mass spectroscopy and ELISA, for nearly 6,000 reagents
SOMAmer Reagent affinity and specificity
demonstrated by multiple methods
SomaLogic provides the largest number of protein measurements and the greatest number of orthogonally confirmed protein reagents in the proteomics industry.
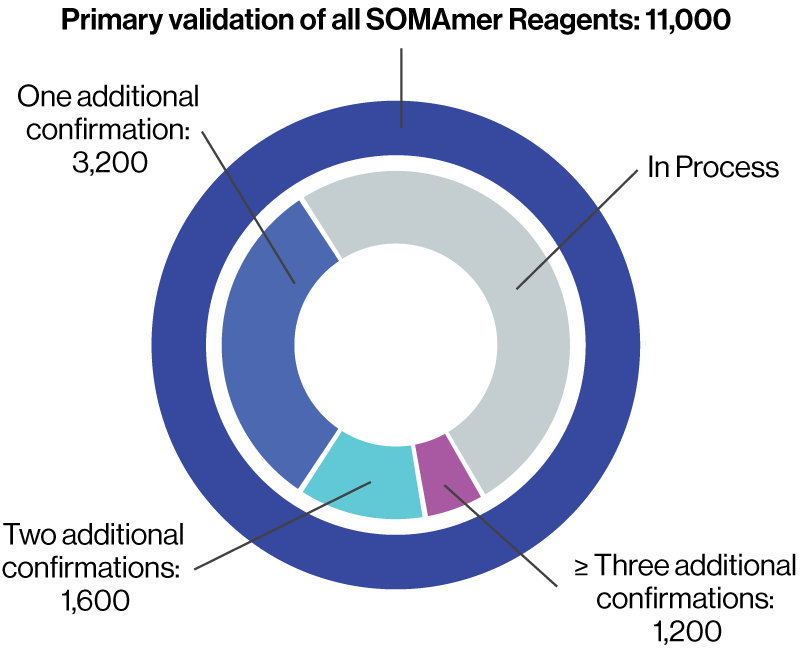
Primary validation of 11,000
SOMAmer Reagents
- Determination of equilibrium binding affinity dissociation constant (KD)
- Pulldown assay of cognate protein from buffer
- Demonstration of buffer dose response in the SomaScan Assay
- Estimation of endogenous cognate protein signals in plasma
Over 6,000 SOMAmer Reagents have at least one additional form of orthogonal confirmation.
Approximately 3,000 SOMAmer Reagents have undergone two or more forms of orthogonal confirmation.




