Why harness the power of proteomics?
When you can profile 11,000 protein measurements from a 55-µL sample with high throughput and reproducibility, you can uncover key insights into disease pathways, pinpoint new treatment targets, and make smarter decisions in your research.
Same genotype. Different phenotype.

Protein assays complement genomics to identify:
- Patient subpopulations
- Novel therapeutic targets
- New disease applications for approved drugs
- Possible safety concerns
- Mechanisms of action
Measure what matters most
Proteins signify health
The ~20,000 proteins that are genetically encoded by the human genome orchestrate the majority of biological processes. By measuring proteins, we can identify patterns of wellness, fitness, and aging.
Proteins reflect treatment
Medications and lifestyle changes cause shifts in the proteome. By measuring proteins, we can determine if treatments are working.
Proteins reveal
risk
Adverse reactions to drugs, health complications, and comorbidities are also determined by proteins. By measuring proteins, we can identify good candidates for clinical trials.
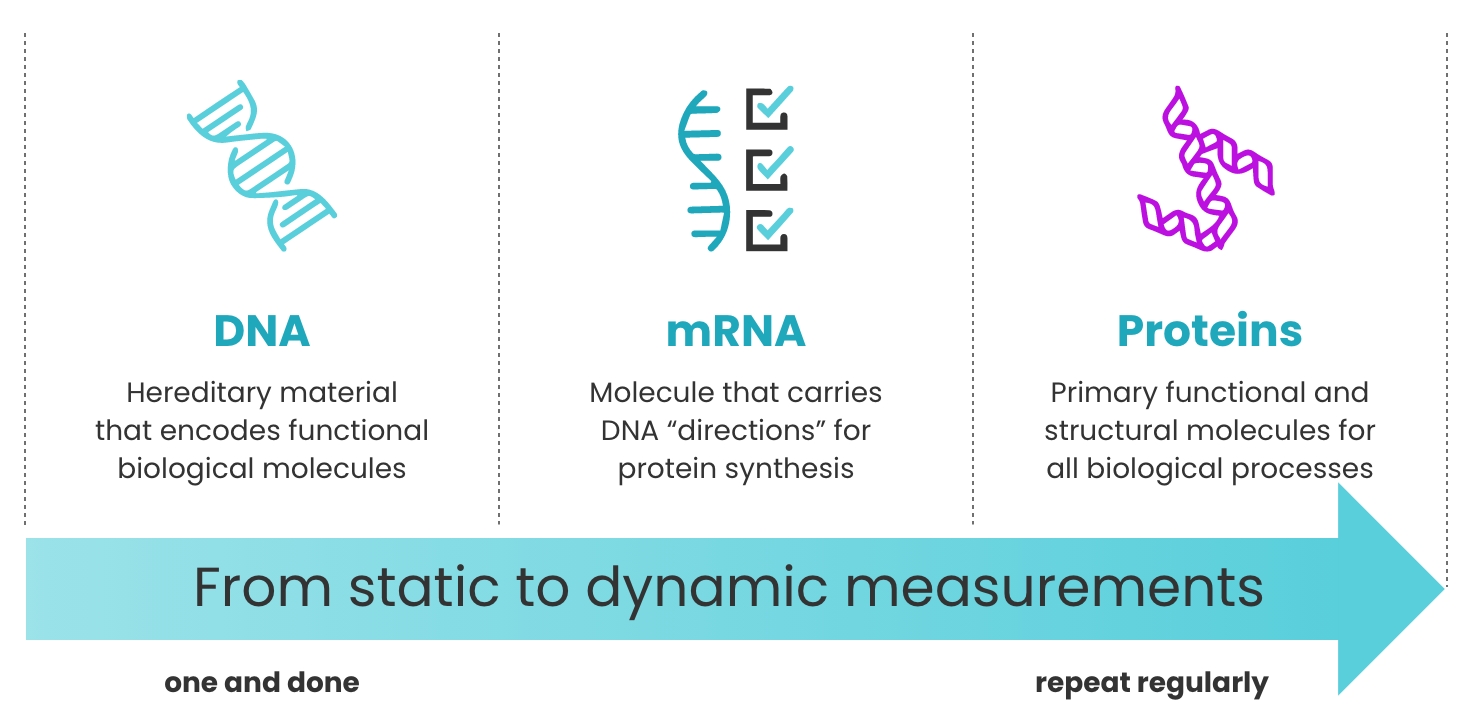
Proteins are dynamic
Unlike the genome, which is relatively fixed over time, proteins provide a snapshot of health in real time.
Proteins are accessible
Key proteins circulate in blood and other fluids, making them easy to sample.
Proteins are the future
In the past, we’ve looked to the genome, because measuring proteins was too technically challenging. With our pioneering platform, we can now measure thousands of proteins across a 10-log range of concentrations from a single sample.




